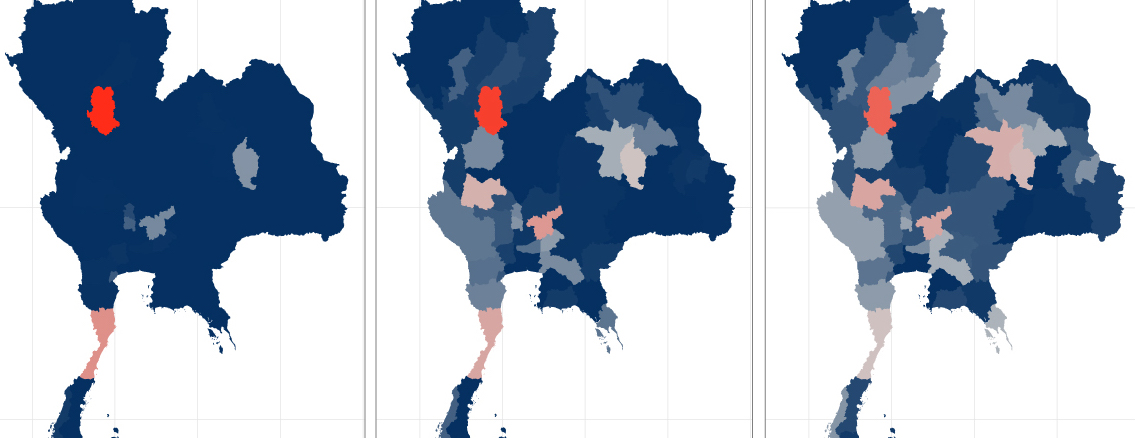
Our team is working to develop statistical methods and tools that can improve real-time infectious disease forecasting efforts for a variety of diseases, including COVID-19, dengue fever and influenza. We focus on building robust and interpretable models that can be used by public health officials to drive policy decisions. For example, in a collaboration with the Ministry of Public Health in Thailand, our team built statistical forecast models to predict outbreaks of dengue fever in real-time from early 2014 through 2019 for each of the 77 provinces in Thailand. We also have worked closely with the CDC to produce ensemble forecasts of influenza and COVID-19. From a methodological perspective we work on developing ensemble methodology (fusing together forecasts from multiple models) and on forecasting methods that can account for reporting delays in surveillance data.
The hubverse is a collection of open-source software and data tools that enables collaborative modeling exercises. It is developed by the Consortium of Infectious Disease Modeling Hubs, a collaboration of research teams and public health professionals that have built and maintained predictive modeling hubs for infectious disease applications. The goal of the Consortium of Infectious Disease Modeling Hubs is to develop a central open-source suite of tools for creating, hosting, maintaining, and running a modeling hub. Working together, we have developed the hubverse for groups that are running collaborative modeling hub efforts.

In multi-pathogen infectious disease systems, complex immunological interactions between multiple strains of disease govern the evolutionary and epidemiological dynamics of disease. Understanding these interactions plays a vital role in clinical and public health decision-making. Our work combines multiple data streams from complex disease systems with modern statistical and computational methodologies to find evidence of complex interactions within the system. For example, our research was the first to use population-level data to explicitly estimate the duration of temporary immunity experienced by individuals after an infection with one of the four serotypes of dengue fever.
We build open-source software motivated by our research projects with the hope that you and others will find these tools useful. From D3 visualization platforms to SQL database utilities to R and python packages, we develop a diverse set of software that help us do better research. In addition to the projects listed above, lab members have also contributed to the development of Stan and the ForecastFramework R package.
Cluster-randomized trials are a type of clinical trial where clusters of individuals are randomized instead of individuals. In our work on several high-profile cluster-randomized clinical trials – including the ResPECT Study (funded by CDC and VA), the SCRUB Trial, and a telehealth collaborative care trial for HIV patients (funded by VA) – we have developed simulation methods for calculating power for cluster-randomized and cluster-randomized crossover trials. These methods are available as an R software package, clusterPower. In other work, we have developed theory around the covariate-adjusted residual estimator (CARE) and its use in both randomized trials and observational settings to estimate causal effects of interest.