Our team develops statistical methods and analytical tools to help people make sense of data. Building on close collaborative relationships with public health practitioners from across the world -- from Denver to San Juan to Bangkok -- we use modern statistical and machine learning tools to gain insights into complex disease systems.
We build software packages, maintain code repositories, develop databases, create interactive data visualizations, and run computationally intensive simulation studies. Depending on the project, we code using mix of R, python, javascript, d3, C, and C++.
Check out our work on GitHub.
Our team develops models for understanding complex and dynamic systems of infectious disease. We have developed real-time forecasts of dengue fever in Thailand, estimated the duration of cross-protection between serotypes of dengue, and predicted the trajectory of the flu season in the US.
Posted on 15 February, 2022
SARS-CoV-2 is the name of the virus that causes COVID-19. There have been a lot of really interesting analyses of SARS-CoV-2 genomic data that look at patterns over time in the circulation of different variants. When we started writing this post, we didn’t know how to use or obtain these data, many of which are publicly available. So we wrote this primer for accessing genomic data on SARS-CoV-2 that are available for the US. These data are made public through GenBank and are pre-processed and served publicly by the Nextstrain team. We hope you find it useful!
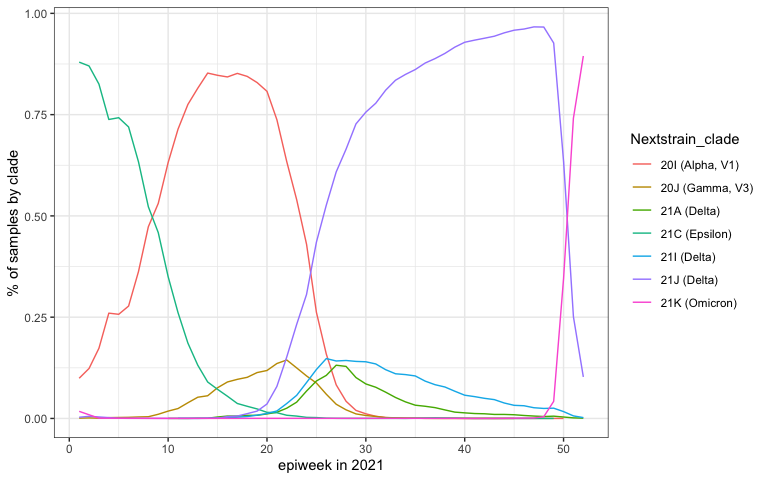
Posted on 17 June, 2019
As I’ve been writing up a progress report for my NIGMS R35 MIRA award, I’ve been reminded at how much of the work that we’ve been doing is focused on forecasting infrastructure. A common theme in the Reich Lab is making operational forecasts of infectious disease outbreaks. The operational aspect means that we focus on everything from developing and adapting statistical methods to be used in forecasting applications to thinking about the data science toolkit that you need to store, evaluate, and visualize forecasts. To that end, in addition to working closely with the CDC in their FluSight initiative, we’ve been doing a lot of collaborative work on new R packages and data repositories that I hope will be useful beyond the confines of our lab. Some of these projects are fully operational, used in our production flu forecasts for CDC, and some have even gone through a level of code peer review. Others are in earlier stages of development. My hope is that in putting this list out there (see below the fold) we will generate some interest (and possibly find some new open-source collaborators) for these projects.
Posted on 29 January, 2019
Want to learn how to do some forecasting with R? Here’s your chance to try out a new time-series forecasting package for R whose aim is to standardize and simplify the process of making and evaluating forecasts!
The Reich Lab uses an R package called ForecastFramework to implement forecasting models. There are many benefits to using ForecastFramework in a forecasting pipeline, including: standardized and simplified rapid model development and performance evaluation. ForecastFramework was created by Joshua Kaminsky of the Infectious Disease Dynamics Group at Johns Hopkins University. The package is open source and can be found on Github.
After watching students in the lab working on learning how to use ForecastFramework, I decided to create a step-by-step demonstration of the primary use cases of ForecastFramework. The complete demo lives at reichlab.io/forecast-framework-demos/.
Posted on 17 September, 2018
Last week, I attended a Pandemic Influenza Exercise at the US CDC. To be clear, there is NOT a pandemic occuring right now, but the CDC ran this exercise where hundreds of staff members and outside observers and participants came together to practice going through the motions of a public health response to a major pandemic. As someone who is usually sheltered from this everyday aspect of public health decision-making, this was a fascinating window into understanding the careful, if time-pressured, scientific deliberation that underlies the response to public health emergencies.

Posted on 28 November, 2017
In March 2017, a group of influenza forecasters who have participated in the CDC FluSight challenge in past seasons established the FluSight Network, a multi-institution and multi-disciplinary consortium of forecasting teams. This group worked throughout 2017 to create a public, real-time collaborative ensemble forecasting model that provides updated forecasts of influenza in the US each week.